Présentation
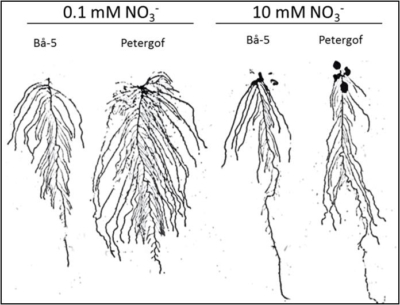
Nitrogen (N) is the quantitatively most important nutrient required by crops, however a considerable fraction of N fertilizer gets lost as runoffs with detrimental consequences to the environment. Faced with those pressing societal costs, innovation has to make a step change to improve nitrogen use efficiency of plants. While the root organ is largely out of sight, we aim at modifying root architecture (more branched system) to take up N sources in the soil more efficiently. The goal of our research is to first identify the genetic determinism that regulate N-related root morphology in a model species (Arabidopsis), then to use that information in a parented crop (oilseed rape) for redesigning its root architecture. In this project, naturally occurring genetic variability is studied in a diversity panel of 360 Arabidopsis populations collected around the world.
The ULB partner is carrying a genome-wide association mapping strategy, in order to identify key genes that determine root morphology in response to nitrate supply. The Cambridge partner is providing a combination of unique genetic and microscopy techniques in concert with advanced 3D computer visualization methods for characterizing the role played by those identified genes in shaping root architecture. Two seminars and short stay visits by the principal investigators in each other institutions and the training of J. De Pessemier (FRIA PhD Student) for 3 months in Cambridge are foreseen during the two years project.
Les promoteurs
- Christian Hermans, Laboratoire de Physiologie et de Génétique moléculaire des Plantes, ULB
- Jim Haseloff, Department of Plant Sciences, Université de Cambridge
Documents
- Rapport final


